Publications (*corresponding author) Google Scholar citations
Priprints
- Busta L.,*, Hall D., Johnson B., Schaut M., Hanson. C.M., Gupta A., Gundrum M., Wang Y., Maeda H.A.* (2024) Phylochemical mapping of natural products onto the plant tree of life using text mining and large language models. bioRvix doi: 10.1101/2024.02.16.580694
Published articles
- *Koper K.#, Han S-W.#, Kothadia R., Salamon H., Yoshikuni Y.*, Maeda H.A.* (2024) Multi-substrate specificity shaped the complex evolution of the aminotransferase family across the tree of life. Proc. Natl. Acad. Sci. 121, e2405524121 bioRvix doi: 10.1101/2024.03.19.585368 #These authors contributed equally.
- Jung S., Maeda H.A.* (2024) Debottlenecking the DOPA 4,5-dioxygenase step with enhanced tyrosine supply boosts betalain production in Nicotiana benthamiana. Plant Physiol. kiae166
- Synthetic biology provides emerging tools to produce valuable compounds in plant hosts as sustainable chemical production platforms. By testing a series of synthetic constructs for biosynthesis of betalain pigments from L-tyrosine, this study revealed that balancing between enhanced supply (“push”) and effective utilization (“pull”) of precursor by alleviating a bottleneck step is critical in producing high levels of target compounds.
- Maeda H.A.*, de Oliveira M.V.V. (2024) VAS1-mediated nitrogen shufffling for aromatic amino acid homeostasis. Trends Plant Sci. 29, 507-509
- El-Azaz J., Moore B., Takeda-Kimura Y., Yokoyama R., Wijesingha Ahchige M., Chen X., Schneider M., Maeda H.A.* (2023) Coordinated regulation of the entry and exit steps of aromatic amino acid biosynthesis supports the dual lignin pathway in grasses. Nature Commun. 14, 7242
- Plants harness sunlight to generate thousands of chemical compounds from CO2, yet how they control the distribution of high-energy carbon remains poorly understood. This study by El-Azaz et al. revealed precise and coordinated regulation of grass enzymes that ensures balanced production of tyrosine and phenylalanine. This likely enables the robust synthesis of phenylpropanoid compounds like lignin in grasses from these two aromatic amino acids as starting materials. Understanding this process now lets us engineer plants to efficiently produce valuable phenolic compounds from atmospheric CO2.
- De Raad M., Koper K., Deng K., Bowen B.P., Maeda H.A., Northen T.R.* (2023) Mass spectrometry imaging-based assays for aminotransferase activity reveal a broad substrate spectrum for a previously uncharacterized enzyme. J. Biol. Chem. 299, 102939.
- Koper K.*, Hataya S., Hall A.G., Takasuka T.E., Maeda H.A.* (2022) Biochemical characterization of plant aromatic aminotransferases Methods Enzymol. 680, 35-83
- Yokoyama R.#, de Oliveira M.V.V.#, Takeda-Kimura Y., Ishihara H., Alseekh S., Arrivault S., Kukshal V., Jez JM, Stitt M., Fernie A.R., Maeda H.A.* (2022) Point mutations that boost aromatic amino acid production and CO2 assimilation in plants Science Adv. 8, eabo3416, Highlighted in UW News, Wisconsin Public Radio, NSF News, SpectrumNews1. # Two authors contributed equally
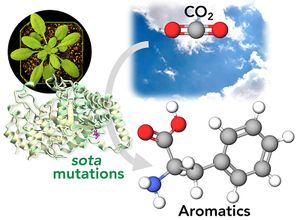 - Here we discovered genetic mutations in plants that enhance the conversion of carbon dioxide (CO2) into aromatic compounds. Aromatics are found everywhere in our society from food ingredients, fuels, plastics, cosmetics, and pharmaceuticals like aspirin. Today, they are mainly produced from fossil fuels. It turns out that plants are very good at producing aromatics, and they do so by using atmospheric CO2 they capture through photosynthesis. In this study, we found that plants have a very sophisticated “brake” at the beginning of the aromatic amino acid pathway that directly connects photosynthesis and synthesis of aromatic compounds in plants. Through genetic screening using Arabidopsis thaliana plants, we identified a series of point mutations, called sota mutations, that can release this brake. Now these plants produce a lot of aromatic amino acids, providing exciting opportunities to achieve sustainable production of aromatic compounds using plants while reducing atmospheric CO2 at the same time.
- Huß S., Judd R.S., Koper K., Maeda H.A., Nikoloski Z.*(2022) An automated workflow that generates atom mappings for large scale metabolic models and its application to Arabidopsis thaliana Plant J. 111, 1486-1500
- Koper K., Han S-W., Pastor D.C., Yoshikuni Y., Maeda H.A.* (2022) Evolutionary Origin and Functional Diversification of Aminotransferases J. Biol. Chem. 298: 102122'
- Yokoyama R.*, Kleven B., Gupta A., Wang Y., Maeda H.A.* (2022) 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase as the gatekeeper of plant aromatic natural product biosynthesis (2022) Curr. Opin. Plant Biol. 67:102219
- Lopez-Nieves S., El-Azaz J., Men Y., Holland C.K., Feng T., Brockington S.F., Jez J.M., Maeda HA* (2022) Two independently evolved natural mutations additively deregulate TyrA enzymes and boost tyrosine production in planta. Plant J 109, 844-855 (full article)
- Tyrosine is a critical amino acid precursor for biosynthesis of numerous plant natural products, including betalain pigments that are uniquely produced in the plant order Caryophyllales. This study identified key residues involved in the evolution of deregulated TyrA enzymes that underlie the overaccumulation of tyrosine and betalains in core Caryophyllales. Furthermore, combined introduction of the identified residue together with one independently evolved in legume TyrAs (Schenck et al., 2017) converted highly-regulated Arabidopsis TyrA into a tyrosine-insensitive enzyme, demonstrating the utility of these natural mutations in elevating tyrosine levels in plants.
- Maeda H.A.* and Fernie A.R.* (2021) Evolutionary History of Plant Metabolism Ann. Rev. Plant Biol. 72, 185-216
- Yokoyama R., de Oliveira V.V.M., Kleven B., Maeda H.A.* (2021) The Entry Reaction of the Plant Shikimate Pathway is Subjected to Highly Complex Metabolite-Mediated Regulation
Plant Cell 33, 671–696
- The shikimate pathway interconnects the central carbon metabolism and biosynthesis of aromatic amino acids and a wide range of aromatic natural products. This study found that the first enzyme of the shikimate pathway, DAHP synthases (DHSs), are regulated by at least five downstream metabolites (i.e. tyrosine, tryptophan, chorimate, arogenate, caffeate), uncovering much more complex feedback regulation of the shikimate pathway in plants than in microbes. The findings will help us enhance the production of these essential amino acids and natural products in plants.
- Zhu F, Alseekh S, Koper K, Tong H, Nikoloski Z, Naake T, Liu H, Yan J, Brotman Y, Wen W, Maeda H, Cheng Y, Fernie AR*. (2021) Genome-wide association of the metabolic shifts underpinning dark-induced senescence in Arabidopsis. Plant Cell online
- Gibbs NM, Su SH, Lopez-Nieves S, Mann S, Alban C, Maeda HA, Masson PH*. (2021) Cadaverine regulates biotin synthesis to modulate primary root growth in Arabidopsis. Plant J 107:1283-1298.
- Yoo H, Shrivastava S, Lynch JH, Huang XQ, Widhalm JR, Guo L, Carter BC, Qian Y, Maeda HA, Ogas JP, Morgan JA, Marshall-Colón A, Dudareva N*. Overexpression of arogenate dehydratase reveals an upstream point of metabolic control in phenylalanine biosynthesis. Plant J 2021, 108:737-751.
- Naake T., Maeda H.A., Proost S., Tohge T., Fernie A.R.* (2020) Kingdom-wide analysis of the evolution of the plant type III polyketide synthase superfamily. Plant Physiol. online
- Schenck C.A., Westphal J., Jayaraman D., Garcia K., Wen J., Mysore K.S., Ané J.M., Sumner L.W., Maeda H.A.* (2020) Role of Cytosolic, Tyrosine-Insensitive Prephenate Dehydrogenase in Medicago truncatula. Plant Direct 4: e00218
- Maeda H.A.* (2019b) Harnessing evolutionary diversification of primary metabolism for plant synthetic biology J. Biol. Chem., 294, 16549-16566.' Invited review in "Natural product biosynthesis: What's next? A thematic series"
- Maeda H.A.* (2019a) Evolutionary diversification of primary metabolism and its contribution to plant chemical diversity Front. Plant Sci., 10 July 2019 Invited review on the special issue, "The Origin of Plant Chemodiversity".
- Wang M., Toda K., Block A., Maeda H.A.* (2019) TAT1 and TAT2 tyrosine aminotransferases have both distinct and shared functions in tyrosine metabolism and degradation in Arabidopsis thaliana. J. Biol. Chem. 294, 3563-3576.
- Lopez-Nieves S.*, Pringle A., Maeda H.A. (2019) Biochemical characterization of TyrA dehydrogenases from Saccharomyces cerevisiae (Ascomycota) and Pleurotus ostreatus (Basidiomycota) Arch Biochem Biophys. [Epub ahead of print]
- de Oliveira M.V.V., Jin X., Chen X., Griffith D., Batchu S., Maeda H.A.* (2019). Imbalance of tyrosine by modulating TyrA arogenate dehydrogenases impacts growth and development of Arabidopsis thaliana.Plant J. 97, 901-922.
- Smith S.D.*, Angelovici R., Heyduk K., Maeda H.A., Moghe G.D., Pires J.C., Widhalm J.R., Wisecaver J.H. (2019). The Renaissance of Comparative Biochemistry. Am. J. Bot.. 106, 3-13.
- Lopez-Nieves S., Yang Y., Timoneda T., Wang M., Feng T., Smith S.A., Brockington S.F., Maeda H.A.* (2018) Relaxation of Tyrosine Pathway Regulation Underlies the Evolution of Betalain Pigmentation in Caryophyllales. New Phytologist 217, 896-908. Free view-only version Featured in Nature Plants, UW News, USDA NIFA, NY Times, and BBC (Spanish)
- Timoneda A., Sheehan H., Feng T., Lopez-Nieves S., Maeda H.A., Brockington S. (2018) Redirecting Primary Metabolism to Boost Production of Tyrosine-Derived Specialised Metabolites in Planta. Sci. Rep. 8; 17256
- Schenck C.A.. and Maeda H.A.* (2018) Tyrosine Biosynthesis, Metabolism, Catabolism in Plants. Phytochemistry 149, 82-102.
- This article provides a comprehensive review on tyrosine biosynthesis, catabolism, and metabolism to downstream specialized metabolites in plants.
- Hollland CK, Berkovich DA, Kohn ML, Maeda H, Jez JM. (2018) Structural Basis for Substrate Recognition and Inhibition of Prephenate Aminotransferase from
Arabidopsis. Plant J. doi: 10.1111/tpj.13856.
- Schenck C.A., Men Y. and Maeda H.A.* (2017b) Conserved Molecular Mechanism of TyrA Dehydrogenase Substrate Specificity Underlying Alternative Tyrosine Biosynthetic Pathways in Plants and Microbes Frontiers Mol. Biosci. online
- Schenck C.A., Holland C.K., Schneider M., Men Y., Lee S.G., Jez J. and Maeda H.A.* (2017a) Molecular Basis of the Evolution of Alternative Tyrosine Biosynthetic Routes in Plants. Nature Chem. Biol. 13, 1029-1035 Free view-only version
Featured in UW News and Nature Plants.
- Wang M. and Maeda H.A.* (2017) Aromatic Amino Acid Aminotransferases in Plants. Phytochemistry Reviews, 1-29. DOI:10.1007/s11101-017-9520-6. Free view-only version
- Lynch JH, Orlova I, Zhao C, Guo L, Jaini R, Maeda H, Akhtar T, Cruz-Lebron J, Rhodes D, Morgan J, Pilot G, Pichersky E, Dudareva N. (2017) Multifaceted plant
responses to circumvent Phe hyperaccumulation by downregulation of flux through the shikimate pathway and by vacuolar Phe sequestration. Plant J. 92, 939-950.
- Wang M., Lopez-Nieves S., Goldman I., Maeda H.A.* (2017) Limited Tyrosine Utilization Explains Lower Betalain Contents in Yellow than Red Table Beet Genotypes. J Agric Food Chem. 65, 4305–4313.
- Betalains are red and yellow alkaloid pigments derived from tyrosine. Previous studies showed that yellow pigments accumulate much less than red pigments. Through quantitative chemical analyses of different beet genotypes, this study found that increased tyrosine levels were positively correlated with elevated betalain accumulations among red but not yellow genotypes, suggesting that yellow beets are not efficiently converting tyrosine into betalain pigments. Thus, better utilization of the accumulated tyrosine can likely improve betaxanthin production in yellow beets.
- Wang M., Toda K., Maeda H.A.* (2016) Biochemical Properties and Subcellular Localization of Tyrosine Aminotransferases from Arabidopsis thaliana. Phytochemistry, 132, 16–25
- Maeda H.A.* (2016) Lignin biosynthesis: Tyrosine shortcut in grasses. Nature Plants 2, 16080 DOI: 10.1038/NPLANTS.2016.80
-
- Schenck C.A., Chen S., Siehl D., Maeda H.A.* (2015) Non-plastidic, Tyrosine-Insensitive Prephenate Dehydrogenases from Legumes. Nature Chem. Biol. 11, 52-57 *Featured on the Cover.
- L-Tyrosine (Tyr) and its plant-derived natural products are essential in both plants and humans. In plants, Tyr is generally assumed to be synthesized in the plastids via arogenate dehydrogenase (ADH) that is strictly inhibited by Tyr. Here we identified non-plastidic Tyr-insensitive prephenate dehydrogenases (PDHs) uniquely present in legumes, providing molecular evidence for the diversification of primary metabolic Tyr pathway via an alternative cytosolic PDH pathway in plants. Also, the Tyr-insensitive property of the identified PDH enzymes has immediate impacts on metabolic engineering to improve the production of Tyr and Tyr-derived natural products.
- Dornfeld C., Weisberg A.J., Ritesh KC, Dudareva N., Jelesko J.G., Maeda H.A.* (2014) Phylobiochemical Characterization of Class-Ib Aspartate/Prephenate Aminotransferases Reveals Evolution of the Plant Arogenate Phenylalanine Pathway Plant Cell 26, 3101-3114
- Plants use phenylalanine to produce abundant and diverse phenylpropanoid compounds, such as flavonoids, tannins, and lignin. Through phylogenetic, bioinformatic, and biochemical analyses of prephenate aminotransferase enzymes from plant and bacterial lineages, this study revealed unique evolutionary history and molecular changes of key enzymes responsible for phenylalanine biosynthesis in plants. The findings assist the rational design of antimicrobial drugs and herbicides, but also highlight the use of phylobiochemical characterization of enzymes from deep taxonomic lineages in determining key molecular changes that lead to the evolution of new metabolic pathways. UW news release.
- Luby C., Maeda H.A., Goldman I. (2014) Genetic and Phenological Variation of Tocochromanol (Vitamin E) Content in Wild (Daucus carota L. var. carota) and Domesticated Carrot (D. carota L. var. sativa) Horticulture Research 1:15
- Maeda H., Song W., Sage T.L., DellaPenna D. (2014) Role of Callose Synthases in Transfer Cell Wall Development in Tocopherol Deficient Arabidopsis Mutants. Front. Plant Sci. 5:46.
- Yoo H., Widhalma J.R., Qiana Y., Maeda H., Cooperc B.R., Jannaschc A.S., Gondae I., Lewinsohne E., Rhodes D., Dudareva D. (2013) An Alternative Pathway Contributes to Phenylalanine Biosynthesis in Plants via a Cytosolic Tyrosine:Phenylpyruvate Aminotransferase. Nature Commun. 4:2833
- Maeda H. and Dudareva N. (2012) The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Ann. Rev. Plant Biol. Vol. 63, 73-105
- Muhlemann J.K., Maeda H., Chang C.Y., San Miguel P., Baxter I., Cooper B., Perera M.A., Nikolau B.J., Vitek O., Morgan J.A., Dudareva N. (2012) Developmental Changes in the Metabolic Network of Snapdragon Flowers. PLoS ONE 7(7): e40381
- Maeda H., Yoo H., and Dudareva N. (2011) Prephenate Aminotransferase Directs Plant Phenylalanine Biosynthesis via Arogenate. Nature Chem. Biol., DOI:10.1038/nchembio.485
- Maeda H., Shasany A.K., Schnepp J., Orlova1 I., Taguchi G., Cooper B.R., Rhodes D., Pichersky E. and Dudareva N. (2010) RNAi Suppression of Arogenate Dehydratase1 Reveals That Phenylalanine Is Synthesized Predominantly via the Arogenate Pathway in Petunia Petals. Plant Cell 22, 832-849 *Described as a Research Highlight in Nature Chem. Biol. 6, 310
- Song W., Maeda H., and DellaPenna D. (2010) Mutations of the ER to plastid lipid transporters (TGD1, 2, 3 and 4) and the ER oleate desaturase (FAD2) suppress the low temperature-induced phenotype of Arabidopsis tocopherol deficient mutant vte2. Plant J. 62, 1004-1018
- Orlova I., Nagegowda D.A., Kish C.M., Gutensohn M., Maeda H., Varbanova M., Fridman E., Yamaguchi S., Hanada A., Kamiya Y., Krichevsky A., Citovsky V., Pichersky E., and Dudareva N. (2009) The Small Subunit Snapdragon Geranyl Diphosphate Synthase Modifies the Chain Length Specificity of Tobacco Geranylgeranyl Diphosphate Synthase in Planta. Plant Cell 21, 4002-4017
- Maeda H., Sage T.L., Isaac G.., Welti R., and DellaPenna D. (2008) Tocopherols Modulate Extra-Plastidic Polyunsaturated Fatty Acid Metabolism in Arabidopsis at Low Temperature. Plant Cell 20, 452-470 *Described in the Featured Article of the issue Plant Cell 20, 246
- Maeda H. and DellaPenna D. (2007) Tocopherol Functions in Photosynthetic Organisms. Curr. Opin. Plant Biol. 10, 260-265
- Maeda H., Song W., Sage T.L. and DellaPenna D. (2007) Tocopherols Play a Limited Role in Photoprotection but a Crucial Role in Chilling Adaptation in Arabidopsis Leaves. In Current Advances in the Biochemistry and Cell Biology of Plant Lipids, C. Benning and J. Ohlrogge, eds (Aardvark Global Publishing Company, LLC, Salt Lake City, UT), pp. 112-115 PDF download (4.5 MB)
- Maeda H., Song W., Sage T.L. and DellaPenna D. (2006) Tocopherols Play a Crucial Role in Low Temperature Adaptation and Phloem Loading in Arabidopsis. Plant Cell 18, 2710-2732 *Highlighted on the Cover of the issue.
- Sakuragi Y., Maeda H., DellaPenna D. and Bryant D.A. (2006) α-Tocopherol Plays a Role in Photosynthesis and Macronutrient Homeostasis of the Cyanobacterium Synechocystis sp. PCC 6803 That is Independent of its Antioxidant Function. Plant Physiol. 141, 508-521
- Maeda H., Sakuragi Y., Bryant D.A., and DellaPenna D. (2005) Tocopherols Protect Synechocystis sp. Strain PCC 6803 from Lipid Peroxidation. Plant Physiol. 138, 1422-1435
- Cheng Z., Sattler S., Maeda H., Sakuragi Y., Bryant D.A., and DellaPenna D. (2003) Highly Divergent Methyltransferases Catalyze a Conserved Reaction in Tocopherol and Plastoquinone Synthesis in Cyanobacteria and Photosynthetic Eukaryotes. Plant Cell 15, 2343-2356
- Okazawa A., Maeda H., Fukusaki E., Katakura Y., and Kobayashi A. (2000) In Vitro Selection of Hematoporphyrin Binding DNA Aptamers. Bioorg. Med. Chem. Lett. 10, 2653-2656
- Fukusaki E., Kato T., Maeda H., Kawazoe N., Ito Y., Okazawa A., Kajiyama S. and Kobayashi A. (2000) DNA Aptamers that Bind to Chitin. Bioorg. Med. Chem. Lett. 10, 423-425
</ol>
|