Difference between revisions of "Maeda Lab:Publications"
From Maeda Lab
| (28 intermediate revisions by the same user not shown) | |||
| Line 17: | Line 17: | ||
<h3><font>Publications</font> ('''*'''corresponding author)</h3> '''[http://scholar.google.com/citations?user=uQBb60kAAAAJ Google Scholar citations]''' | <h3><font>Publications</font> ('''*'''corresponding author)</h3> '''[http://scholar.google.com/citations?user=uQBb60kAAAAJ Google Scholar citations]''' | ||
| + | '''Priprints''' | ||
| + | *'''Takeda-Kimura Y.''', '''Moore B.''', Holden S., Deb S.K., Barrett M., Lorence D., '''de Oliveira M.V.V.''', Grimwood J., Williams M., Boston L.B., Jenkins J.W., Plott C., Shu S., Barry K.W., Goodstein D.M., Schmutz J., Moscou M.J., McKain M.R., Leebens-Mack J.H.*, '''Maeda H.A.''' (2024) Genomes of Poaceae sisters reveal key metabolic innovations preceding the emergence of grasses. [https://doi.org/10.1101/2024.11.06.622220 '''''bioRvix''''' https://doi.org/10.1101/2024.11.06.622220] | ||
| + | |||
| + | *'''Yokoyama R.*''', '''Maeda H.A.*''' (2024) Arabidopsis 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthases of the shikimate pathway display both manganese- and cobalt-dependent activities. [https://doi.org/10.1101/2024.10.06.616849 '''''bioRvix''''' https://doi.org/10.1101/2024.10.06.616849] | ||
| + | |||
| + | '''Published articles''' | ||
<ol> | <ol> | ||
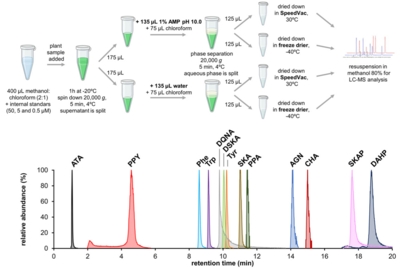
| − | <li>De Raad M., '''Koper K.''', Deng K., Bowen B.P., '''Maeda H.A.''', Northen T.R.*(2023) Mass spectrometry imaging-based assays for aminotransferase activity reveal a broad substrate spectrum for a previously uncharacterized enzyme [https:// | + | <li>'''El-Azaz J.*''', '''Maeda H.A.*''' (2024) A simplified liquid chromatography‐mass spectrometry methodology to probe the shikimate and aromatic amino acid biosynthetic pathways in plants [http://doi.org/10.1111/tpj.17105 '''''Plant J. online'''''] |
| − | <li> '''Koper K.*''', Hataya S., '''Hall A.G.''', Takasuka T.E., '''Maeda H.A.*''' (2022) Biochemical characterization of plant aromatic aminotransferases [https://www.sciencedirect.com/science/article/abs/pii/S0076687922002981?via%3Dihub '''''Methods Enzymol'''''] | + | [[File:FigJorge2024.png|center|400px|]] |
| + | |||
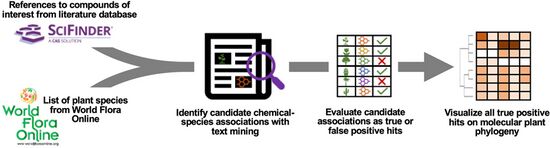
| + | <li>Busta L.,*, '''Hall D.''', Johnson B., '''Schaut M.''', '''Hanson. C.M.''', '''Gupta A.''', '''Gundrum M.''', '''Wang Y.''', '''Maeda H.A.*''' (2024) Phylochemical mapping of natural products onto the plant tree of life using text mining and large language models. [https://doi.org/10.1101/2024.02.16.580694 '''''bioRvix''''' doi: 10.1101/2024.02.16.580694]Now it is published at [http://doi.org/10.1111/tpj.16906 '''''Plant J. online''''']! | ||
| + | |||
| + | [[File:phylochemical1.jpg|center|550px|]] | ||
| + | |||
| + | |||
| + | [[File:PNAS24.jpg|right|300px|]] | ||
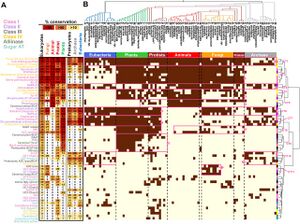
| + | <li>'''Koper K.'''#, Han S-W.#, Kothadia R., Salamon H., Yoshikuni Y.*, '''Maeda H.A.*''' (2024) Multi-substrate specificity shaped the complex evolution of the aminotransferase family across the tree of life. [https://pubmed.ncbi.nlm.nih.gov/38885378/ '''''Proc. Natl. Acad. Sci. 121, e2405524121'''''] [https://www.biorxiv.org/content/10.1101/2024.03.19.585368v1 '''''bioRvix''''' doi: 10.1101/2024.03.19.585368] #These authors contributed equally. | ||
| + | :<font color="#404040"> ''The evolutionary analyses of the aminotransferase (AT) enzyme family across the tree of life found that many essential AT reactions are carried out by nonorthologous enzymes in different taxa, likely due to the multi-substrate specificity exhibited by many AT enzymes. The findings suggest the presence of diverse architecture and functionality of the nitrogen metabolic networks that likely operate in various extant organisms dealing with different nitrogen availabilities and demands.'' | ||
| + | |||
| + | <li>'''Jung S.''', '''Maeda H.A.*''' (2024) Debottlenecking the DOPA 4,5-dioxygenase step with enhanced tyrosine supply boosts betalain production in ''Nicotiana benthamiana''. [https://academic.oup.com/plphys/advance-article-abstract/doi/10.1093/plphys/kiae166/7631229?redirectedFrom=fulltext '''''Plant Physiol. kiae166'''''][[File:DODA.jpg|right|150px|]] | ||
| + | |||
| + | :<font color="#404040"> ''Synthetic biology provides emerging tools to produce valuable compounds in plant hosts as sustainable chemical production platforms. By testing a series of synthetic constructs for biosynthesis of betalain pigments from L-tyrosine, this study revealed that balancing between enhanced supply (“push”) and effective utilization (“pull”) of precursor by alleviating a bottleneck step is critical in producing high levels of target compounds.'' | ||
| + | |||
| + | <li>'''Maeda H.A.*''', '''de Oliveira M.V.V.''' (2024) VAS1-mediated nitrogen shufffling for aromatic amino acid homeostasis. [https://pubmed.ncbi.nlm.nih.gov/38480091/ '''''Trends Plant Sci. 29, 507-509'''''] | ||
| + | |||
| + | <li>'''El-Azaz J.''', '''Moore B.''', '''Takeda-Kimura Y.''', '''Yokoyama R.''', '''Wijesingha Ahchige M.''', '''Chen X.''', '''Schneider M.''', '''Maeda H.A.*''' (2023) Coordinated regulation of the entry and exit steps of aromatic amino acid biosynthesis supports the dual lignin pathway in grasses. [https://www.nature.com/articles/s41467-023-42587-7?utm_source=rct_congratemailt&utm_medium=email&utm_campaign=oa_20231109&utm_content=10.1038/s41467-023-42587-7#Sec25 '''''Nature Commun. 14, 7242'''''][[File:NatCom_Fig1.png|right|300px|]] | ||
| + | |||
| + | :<font color="#404040"> ''Plants harness sunlight to generate thousands of chemical compounds from CO2, yet how they control the distribution of high-energy carbon remains poorly understood. This study by El-Azaz et al. revealed precise and coordinated regulation of grass enzymes that ensures balanced production of tyrosine and phenylalanine. This likely enables the robust synthesis of phenylpropanoid compounds like lignin in grasses from these two aromatic amino acids as starting materials. Understanding this process now lets us engineer plants to efficiently produce valuable phenolic compounds from atmospheric CO2.'' | ||
| + | |||
| + | <li>De Raad M., '''Koper K.''', Deng K., Bowen B.P., '''Maeda H.A.''', Northen T.R.* (2023) Mass spectrometry imaging-based assays for aminotransferase activity reveal a broad substrate spectrum for a previously uncharacterized enzyme. [https://pubmed.ncbi.nlm.nih.gov/36702250 '''''J. Biol. Chem. 299, 102939''''']. | ||
| + | <li> '''Koper K.*''', Hataya S., '''Hall A.G.''', Takasuka T.E., '''Maeda H.A.*''' (2022) Biochemical characterization of plant aromatic aminotransferases [https://www.sciencedirect.com/science/article/abs/pii/S0076687922002981?via%3Dihub '''''Methods Enzymol. 680, 35-83'''''] | ||
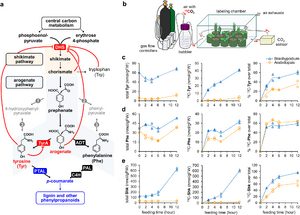
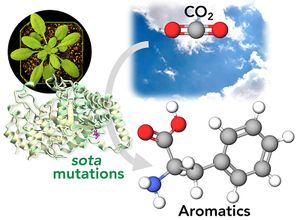
<li>'''Yokoyama R.'''''#'', '''de Oliveira M.V.V.'''''#'', '''Takeda-Kimura Y.''', Ishihara H., Alseekh S., Arrivault S., Kukshal V., Jez JM, Stitt M., Fernie A.R., '''Maeda H.A.*''' (2022) Point mutations that boost aromatic amino acid production and CO2 assimilation in plants [https://www.science.org/doi/10.1126/sciadv.abo3416 '''''Science Adv. 8, eabo3416'''''], ''Highlighted in'' [https://news.wisc.edu/altered-gene-helps-plants-absorb-more-carbon-dioxide-produce-more-useful-compounds/ '''''UW News'''''], [https://www.wpr.org/altered-gene-could-help-plants-absorb-more-carbon-dioxide-study-finds '''''Wisconsin Public Radio'''''], [https://beta.nsf.gov/news/altered-gene-helps-plants-absorb-more-carbon-dioxide-produce-more-useful-compounds '''''NSF News'''''], [https://spectrumnews1.com/wi/milwaukee/news/2022/07/29/madison-scientists-discuss-gene-altering-in-plants- '''''SpectrumNews1''''']. ''# Two authors contributed equally'' [[File:sota_aromatics2.jpg|right|300px|]] | <li>'''Yokoyama R.'''''#'', '''de Oliveira M.V.V.'''''#'', '''Takeda-Kimura Y.''', Ishihara H., Alseekh S., Arrivault S., Kukshal V., Jez JM, Stitt M., Fernie A.R., '''Maeda H.A.*''' (2022) Point mutations that boost aromatic amino acid production and CO2 assimilation in plants [https://www.science.org/doi/10.1126/sciadv.abo3416 '''''Science Adv. 8, eabo3416'''''], ''Highlighted in'' [https://news.wisc.edu/altered-gene-helps-plants-absorb-more-carbon-dioxide-produce-more-useful-compounds/ '''''UW News'''''], [https://www.wpr.org/altered-gene-could-help-plants-absorb-more-carbon-dioxide-study-finds '''''Wisconsin Public Radio'''''], [https://beta.nsf.gov/news/altered-gene-helps-plants-absorb-more-carbon-dioxide-produce-more-useful-compounds '''''NSF News'''''], [https://spectrumnews1.com/wi/milwaukee/news/2022/07/29/madison-scientists-discuss-gene-altering-in-plants- '''''SpectrumNews1''''']. ''# Two authors contributed equally'' [[File:sota_aromatics2.jpg|right|300px|]] | ||
| Line 66: | Line 94: | ||
<li>Timoneda A., Sheehan H., Feng T., '''Lopez-Nieves S.''', '''Maeda H.A.''', Brockington S. (2018) Redirecting Primary Metabolism to Boost Production of Tyrosine-Derived Specialised Metabolites ''in Planta''. [https://www.nature.com/articles/s41598-018-33742-y '''''Sci. Rep. 8; 17256'''''] | <li>Timoneda A., Sheehan H., Feng T., '''Lopez-Nieves S.''', '''Maeda H.A.''', Brockington S. (2018) Redirecting Primary Metabolism to Boost Production of Tyrosine-Derived Specialised Metabolites ''in Planta''. [https://www.nature.com/articles/s41598-018-33742-y '''''Sci. Rep. 8; 17256'''''] | ||
| − | <li>'''Schenck C.A..''' and '''Maeda H.A.*''' (2018) Tyrosine Biosynthesis, Metabolism, Catabolism in Plants. [https:// | + | <li>'''Schenck C.A..''' and '''Maeda H.A.*''' (2018) Tyrosine Biosynthesis, Metabolism, Catabolism in Plants. [https://pubmed.ncbi.nlm.nih.gov/29477627/ '''''Phytochemistry 149, 82-102''''']. |
:<font color="#404040"> ''This article provides a comprehensive review on tyrosine biosynthesis, catabolism, and metabolism to downstream specialized metabolites in plants.'' </font> | :<font color="#404040"> ''This article provides a comprehensive review on tyrosine biosynthesis, catabolism, and metabolism to downstream specialized metabolites in plants.'' </font> | ||
| Line 116: | Line 144: | ||
<li>Okazawa A., '''Maeda H.''', Fukusaki E., Katakura Y., and Kobayashi A. (2000) In Vitro Selection of Hematoporphyrin Binding DNA Aptamers. [http://www.ncbi.nlm.nih.gov/pubmed/11128644 '''''Bioorg. Med. Chem. Lett.''''' 10, 2653-2656]</li> | <li>Okazawa A., '''Maeda H.''', Fukusaki E., Katakura Y., and Kobayashi A. (2000) In Vitro Selection of Hematoporphyrin Binding DNA Aptamers. [http://www.ncbi.nlm.nih.gov/pubmed/11128644 '''''Bioorg. Med. Chem. Lett.''''' 10, 2653-2656]</li> | ||
<li>Fukusaki E., Kato T., '''Maeda H.''', Kawazoe N., Ito Y., Okazawa A., Kajiyama S. and Kobayashi A. (2000) DNA Aptamers that Bind to Chitin. [http://www.ncbi.nlm.nih.gov/pubmed/10743940 '''''Bioorg. Med. Chem. Lett.''''' 10, 423-425]</li> | <li>Fukusaki E., Kato T., '''Maeda H.''', Kawazoe N., Ito Y., Okazawa A., Kajiyama S. and Kobayashi A. (2000) DNA Aptamers that Bind to Chitin. [http://www.ncbi.nlm.nih.gov/pubmed/10743940 '''''Bioorg. Med. Chem. Lett.''''' 10, 423-425]</li> | ||
| + | |||
</ol> | </ol> | ||
Latest revision as of 09:04, 7 November 2024
Publications (*corresponding author)Google Scholar citationsPriprints
Published articles
|